Dr. Jeff Lichtman and colleagues published a paper in Cell outlining a series of techniques for sectioning, reconstructing, and analyzing a discrete volume of mouse brain tissue with nanometer resolution. Advancing the automated collection and processing of cortical tissue at the nanometer scale, this project stems from a collaborative, six-year effort of multiple researchers currently supported by The BRAIN Initiative®.
In collaboration with multiple BRAIN-funded investigators, Dr. Jeff Lichtman of Harvard University leads a recent publication in Cell that describes and tests a new, automated method to probe the physical construction of cortical areas in the brain on a nanometer scale. The approaches and results put forth in the paper directly address multiple goals of the NIH BRAIN Initiative, including cross-disciplinary efforts to develop technologies to better characterize cell types and their connections throughout the brain.
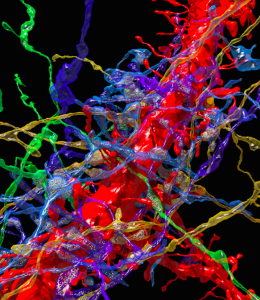
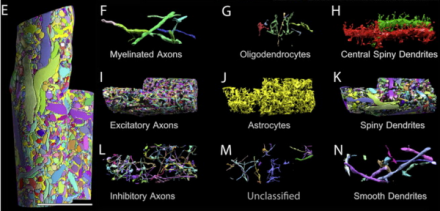
The authors detail an automatic tape-collecting ultra-microtome (ATUM) that uses a diamond knife to slice serial, 29-nanometer sections of mouse brain onto running tape, allowing researchers to accumulate thousands of ultra-fine sections of neocortex. Following imaging of the sections using a scanning electron microscope, the investigators employ a suite of computer programs for virtual reconstruction of the cortical area, and subsequent analysis of cellular processes therein. Investigators analyzed the saturated, reconstructed volume of cortex across multiple scales to identify and quantify cellular objects (such as axons, dendrites, and glial support cells), sub-cellular components (including synapses, synaptic vesicles, and mitochondria), and cellular connectivity. Using a 0.13mm3 volume of mouse somatosensory neocortex, Lichtman and colleagues built a minable database where they could explore differences in physical properties across cells, notably how one might most accurately predict connectivity between excitatory cells. While the authors note numerous analytical challenges presented by the complex, high-resolution data obtained at present, BRAIN Initiative funding across federal agencies incorporates improving methods of analysis and sharing for large datasets – hopefully making such difficulties surmountable in the near future.
Reference: Kasthuri N et al. (2015) Saturated reconstruction of a volume of neocortex. Cell. 162(3):648-661.