New far-red fluorescent protein developed… Improved brain imaging by reducing errors during post-processing… Sublayer-specific coding dynamics in hippocampal pyramidal cells…
Researchers discover a new class of fluorescent protein genes in cyanobacteria and develop a far-red fluorescent protein for potential use in vivo.
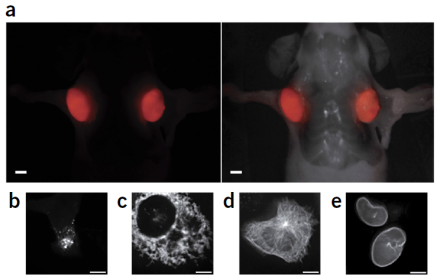
Far-red and near-infrared fluorescent proteins are used for visualizing deep tissue and whole animal in vivo imaging. At present, there are only a few proteins in the far-red spectrum available for use, and the existing red fluorescent proteins generate hydrogen peroxide during fluorescent signal maturation, which can alter immune function or elicit an inflammatory response, making it difficult to study normal tissue function or disease states. To address this limitation, a team of investigators at the University of California at San Diego that includes BRAIN Initiative® grantee Dr. John Y. Lin, isolated a fluorescent protein gene from cyanobacteria and subjected it to a series of mutations to develop a more optimal far-red fluorescent protein. They described these findings in a recent publication in Nature Methods. After 12 rounds of mutagenesis and screening over a million bacterial colonies, they selected and characterized a fluorescent protein with 20 mutations, called small ultra-red fluorescent protein (smURFP). In comparisons with the coral-derived red fluorescent protein mCherry, they found smURFP had comparable expression patterns in neurons and was superior for visualization in deep tissue. The new protein is expressed efficiently, very photo-stable, and biophysically one of the brightest fluorescent proteins created. Additionally, smURFP does not produce hydrogen peroxide, enhancing its potential for studying disease and cell function in vivo. Although the researchers caution that smURFP is not yet ready for use in vivo, smURFP and future derivations of this protein hold great promise for improving the visualization and study of cellular processes in deep tissue, especially in the brains of live animals.
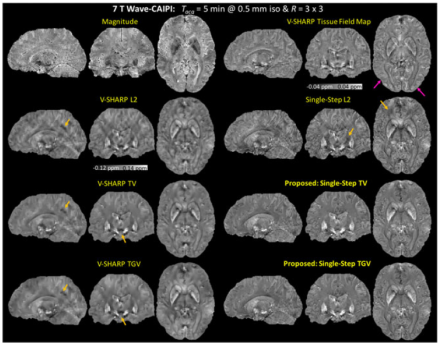
Researchers demonstrate a simplified method of processing MRI images to remove artifacts and reduce image blurring.
Magnetic resonance imaging (MRI) is used to image anatomy and physiological processes in the body. Gradient echo (GRE) is a method of MRI image generation that results in high resolution images, including greater contrast between gray and white brain matter. Quantitative susceptibility mapping (QSM) is a novel technique used to process MRI images and has great potential in studies of brain development and aging. Unfortunately, applying QSM to GRE images requires multiple post-processing steps, which can lead to image blurring and artifacts. In a recent publication in NMR In Biomedicine, a highly collaborative team of researchers at the Massachusetts Institute of Technology, Massachusetts General Hospital, and Harvard Medical School, including BRAIN Initiative grantee Dr. Lawrence L. Wald
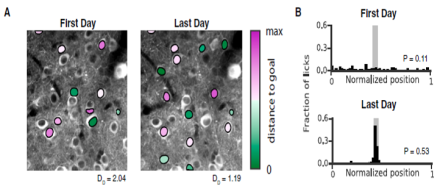
Distinct hippocampal CA1 pyramidal cell subpopulations encode different features of the environment and differentially contribute to learning.
The hippocampus is critical for processing spatial information and episodic memory, and its varied cell types are spatially organized across three axes: dorsal-ventral, proximal-distal, and superficial-deep. Emerging evidence suggests regional functional specialization exists along the first two axes, but specialization along the superficial-deep axis, or any relationship to the dynamics of learning, has yet to be determined. In a recent publication in Neuron, Dr. Atilla Losonczy from Columbia University, who is a BRAIN Initiative co-investigator with Dr. Ivan Soltesz from the University of California (Irvine), and colleagues investigated the hypothesis that distinct hippocampal CA1 pyramidal cell subpopulations encode different features of the environment and differentially contribute to learning. To test this hypothesis, researchers assessed neural activity of two distinct sublayer populations of CA1 pyramidal cells over long time-scales and with subcellular resolution using two-photon calcium activity imaging during two behavioral tasks, random foraging and goal-oriented learning. They found that along the superficial-deep axis, superficial CA1 pyramidal cells provided a more stable place map of the environment during random foraging, while cells in the deep sublayer created a more flexible representation of the environment capable of being shaped by important cues during goal-oriented learning. This work expands our understanding of cell subtypes and their function in the hippocampus, and suggests a mechanism by which the hippocampus can simultaneously convey a stable map of space and process behaviorally-relevant, environmental information for learning.