New classification of channelrhodopsin identified… Novel tool developed for studying cell lineages in whole organisms… Multi-area two-photon microscope monitors cortico-cortical dynamics…
Researchers discover new, functionally similar but structurally different cation channelrhodopsins.
In algae, light-gated ion channels (“channelrhodopsins”) allow differently charged ions to flow across the cell membrane in response to stimulation by light. This discovery led to the development of optogenetics, a powerful tool that uses light to control electrical excitability of cells in living tissue, such as neurons in the brain. Cation channelrhodopsins, originally isolated from chlorophyte (green) algae, evoke light-controlled depolarization of the cell membrane, which can result in activation when expressed in neurons. Anion channelrhodopsins isolated from a distinctly different type of algae, cryptophyte algae, are capable of hyperpolarization of the cell membrane and can be used for neuronal inhibition. In a recent publication in Biophysical Journal, BRAIN Initiative grantee Dr. John Spudich and colleagues at the University of Texas Health Science Center report the discovery of a new set of four different cation channelrhodopsins from cryptophyte algae. The new channelrhodopsins are evolutionarily more closely related to haloarchael proton pumps than to their functionally-related chlorophyte channelrhodopsin counterparts, highlighting the discovery of a convergent process for the evolution of cation channelrhodopsins.
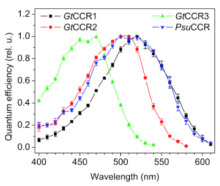
By expressing the four newly discovered channelrhodopsins in HEK293 cells and studying their electrogenic properties via whole-cell patch-clamp recording, the researchers determined that all four channelrhodopsins transport both Na+ and H+ and generate inward currents in response to light, though each has a different wavelength spectrum sensitivity. This discovery expands the repertoire of channelrhodopsins available for manipulation of neural activity, supporting a major goal of The BRAIN Initiative® in developing new and improved technologies for studying the brain.
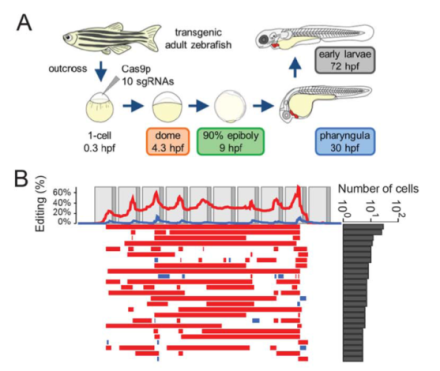
Genome editing of synthetic target arrays for lineage tracing (GESTALT) is a newly developed method for studying cell lineage in complex, whole organisms.
Multicellular organisms are derived from single cells through distinct developmental patterns, or cell lineages. Despite the existence of several approaches for marking cells and tracing cell lineages, investigating large-scale reconstruction of cell lineages in multicellular systems remains a challenge. In a recent Science publication, the lab of Dr. Jay Shendure from the University of Washington, in collaboration with BRAIN Initiative awardee Dr. Alexander F. Schier, has developed genome editing of synthetic target arrays for lineage tracing (GESTALT). This method uses CRISPR/Cas9 genome editing to create an array of target sites called a “barcode”. The barcode marks cells and uses the patterns of mutations shared between cells to determine lineage relationships. The authors applied this method to both artificial lineages (i.e., cell culture using human embryonic kidney 293T cells) and to a whole organism, the zebrafish. Their application of GESTALT suggests that relatively few embryonic progenitor cells give rise to the majority of cells in the organs of the adult zebrafish. The full tree reconstructions for the cell culture, zebrafish embryo, and zebrafish adult experiments can be found here. Although much work needs to be done to optimize GESTALT, this proof-of-principle study facilitates the BRAIN Initiative’s goal of a census of cell types in the brain and reveals a powerful new tool for the field of developmental biology with implications for studying normal development, as well as diseases in which cell division goes wrong, such as cancer, and inherited diseases, such as Down syndrome and cystic fibrosis.
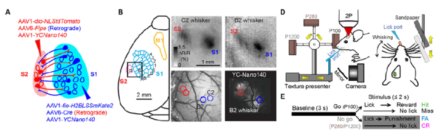
A novel two-photon microscope combined with anatomical tracers simultaneously monitors neural activity across multiple regions of the somatosensory cortex of the awake-behaving mouse.
Coordination of neural activity across multiple brain regions gives rise to behavior and cognitive function. One of the goals of the BRAIN Initiative is to produce a dynamic picture of the functioning brain by developing and applying improved methods for large‐scale monitoring of neural activity – to find a way to visualize the coordination of this activity. Although the advancement of two-photon microscopy using calcium imaging has provided the ability to monitor action potentials across neuronal populations within one cortical area, it has not been possible to image long-range activity across multiple cortical regions. In a recent article in eLife, international BRAIN Initiative grantee Dr. Fritjof Helmchen and colleagues describe a new microscope system that allowed for the simultaneous measurement of activity of individual neurons across the primary (S1) and secondary (S2) somatosensory neocortices of awake-behaving mice. The authors identified coordinated feed-forward and feedback neuronal communication between these cortical regions during a texture discrimination behavioral task. Although both movement and sensory stimulation activated the primary and secondary areas of the mouse whisker cortex, the direct cortico-cortical connections mainly exchanged sensory information. The scientists’ approach using a multi-area two-photon microscope in combination with viral-mediated labeling of long-range projection neurons can be used to help provide a more comprehensive view of how the brain functions under normal conditions, as well as in models of disease states.