Novel neural network translates cortical activity to text… Low-carb diet may prevent and reverse brain aging… Transcriptomic characterization of rare von Economo neurons… Deep learning model predicts Parkinson’s symptomology...
Encoder-decoder framework translates human cortical activity to text
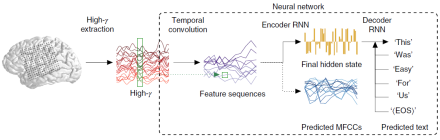
Brain-machine interfaces (BMIs) allow tetraplegics to speak by decoding brain activity into text or speech. Direct decoding of speech, however, is typically restricted to single phonemes or monosyllables, and decoding continuous speech is both limited in vocabulary size and prone to errors. By leveraging advances in machine translation – the computational translation of text from one language to another – Dr. Edward Chang and his research team at the University of California, San Francisco crafted a machine learning framework to translate cortical brain activity to text with high accuracy and at natural-speech rates. Rather than base their decoding on single words, researchers trained a neural network model to translate brain signals from electrocorticograms (ECoGs) into whole written English sentences. In the study, four consenting participants undergoing treatment for epilepsy read a series of sentences displayed on a screen out loud. While they read, researchers recorded their brain activity using ECoG arrays of 120-250 electrodes implanted over each participant’s peri-Sylvian cortex. Then, these data sequences were fed into an ‘encoder-decoder’-style artificial neural network. The network processes the sequences in three stages: (1) it extracts temporal or timing patterns in the signals (i.e., temporal convolution); (2) the data are passed to an encoder recurrent neural network (RNN), which learns to summarize sequences in a single abstract state; and (3) the abstract state is decoded or transformed into written text by a second RNN, which learns to predict the next word in the sequence. For each participant, speech was limited to a restricted ‘language’ of 30-50 sentences. Nevertheless, the framework was highly accurate, as word error rates were as low as three percent on datasets with 250-word vocabularies. Further, speech decoded with minimal data was improved by transfer learning, meaning that the network can potentially learn new English words that it had not trained on from other participants or speaking tasks. The novel framework created by Dr. Chang and his team represents significant progress in using artificial neural networks to decode natural speech and has the potential to improve current speech prosthetics in patients.
***
Neuroimaging study shows that a low-carb diet may prevent, reverse brain aging
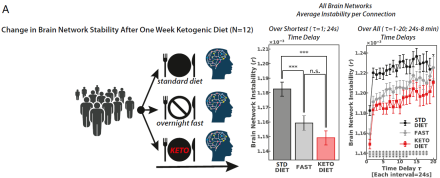
Brain aging, and especially dementia, is associated with hypometabolism – the inability of neurons to use glucose as an energy source. Converging evidence in animal models also suggests that switching the brain’s fuel source from glucose to ketones could restore and maybe even prevent the neurobiological changes associated with cognitive decline. However, little is known about the time course of these age-related changes and how diet influences brain aging in humans. In a recent neuroimaging study, Dr. Lilianne R. Mujica-Parodi and her colleagues at Stony Brook University showed that the neurobiological changes associated with aging are observed at a younger age than previously thought and that this process can be prevented or reversed by a diet low in carbohydrates. First, Dr. Mujica-Parodi and her team used two large-scale human fMRI datasets from 928 individuals across the life span (ages 18 to 88) to establish network stability as a robust whole-brain biomarker associated with aging. They found that functional communication between brain regions destabilizes with age, typically beginning in the late 40s, and that this biomarker is linked to poorer cognition and accelerates insulin resistance. Next, researchers conducted two neuroimaging experiments in 42 adults under the age of 50 to test the effects of manipulating fuel type: glucose versus ketone bodies, on the biomarker. For the first group, each participant as scanned while on a standard diet, overnight fasting, and ketogenic diet. To isolate the effects of fuel type, a second group fasted overnight and was scanned before and after receiving a calorie-matched glucose and ketone ester supplement. Dr. Mujica-Parodi and her team found that brain networks were destabilized by glucose and stabilized by ketones, regardless of whether ketosis was achieved via changes in diet or the calorie-rich supplement. Taken together, these findings suggest that brain network destabilization may reflect early signs of hypometabolism and that simple dietary interventions may protect the aging brain. To learn more about these fascinating results, please read the Stony Brook University news release.
***
Transcriptomics and cross-species homology mapping used to characterize von Economo neurons
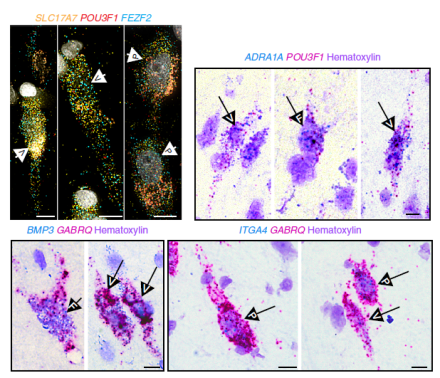
von Economo neurons (VENs) are large, spindle-shaped neurons in the frontal cortex that are particularly vulnerable to neuropsychiatric and neurodegenerative diseases. In humans, these rare neurons only reside in the anterior cingulate or insular cortices and are not found in rodent brains, making their functional properties challenging to study. Recent studies using postmortem brain tissue data from the Allen Human Brain Atlas have identified several genetic markers of VENs. Other studies have identified possible projection targets, but distinct features of VENs remain unknown. To more thoroughly characterize VENs, Dr. Ed Lein and his research team at the Allen Institute for Brain Science used modern RNA-sequencing technologies in human and mouse brains to determine that VENs are a regionally distinct type of extratelencephalic-projecting excitatory neuron. Researchers first used single-nucleus RNA-sequencing (snRNA-seq) to closely examine the genetic profile of cells from the frontoinsular cortex of two postmortem human brain specimens. SnRNA-seq is a valuable tool for classifying and characterizing human brain cells because it can be used on postmortem frozen tissue and it enables the alignment of cell type datasets across brain areas and species. By using this method, researchers identified 13 excitatory neuron types and found that VENs can be localized to a single transcriptomic cell type. This classification included cells with fork and pyramidal morphologies, however, researchers also identified new genetic markers for VENs. Next, to further characterize the cellular properties of VENs, the team carried out cross-species homology mapping by aligning the human snRNA-seq data with mouse cortical single-cell RNA-sequencing data. They found that the human cells were homologous to extratelencephalic (ET) neurons in the mouse, enabling them to predict that, just like mouse ET neurons, human VENs send projections subcortically. Researchers also performed the very first ex vivo electrophysiological recordings of VENs in postmortem human brain tissue and showed that these cells had distinct intrinsic membrane properties of nearby pyramidal neurons. These findings demonstrate how advanced transcriptomic approaches that enable precise cell-type identification and mapping across species enable a better understanding of human brain cells and their circuitry.
***
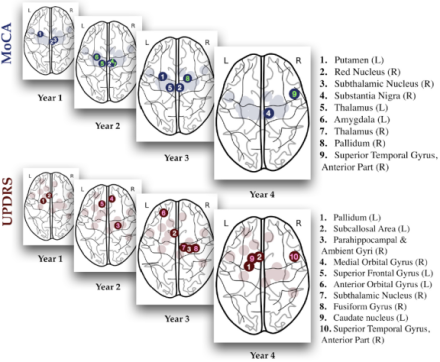
Deep learning neural network uncovers vital role of brain regions in Parkinson’s symptomology
Machine learning (ML) has emerged as a useful tool for neurodegenerative disease diagnosis and progression. Recent ML studies have used deep-learning models to predict Alzheimer’s symptomology and observable cognitive and motor symptoms in Parkinson’s disease (PD) patients. However, the high variability of PD brains, small sample sizes, and lack of long-term brain atrophy make estimating future PD symptomology with deep-learning networks a challenge. In this study, Dr. Ashish Raj and his research team at the University of California, San Francisco employed an autoencoder-based deep learning model to predict future cognitive and motor impairments in PD patients. Researchers selected structural fMRI data from 42 healthy controls and 116 PD patients from the Parkinson’s Progression Markers Initiative database. Using this dataset, they designed a neural network model that predicted a patients’ cognitive and motor score over a few years simply based on anatomical features of brain regions at baseline. Importantly, training and testing the model was quicker and computationally inexpensive compared to existing neural networks that perform similarly. Overall, their model identified salient brain regions involved in cognitive and motor decline, such as the substantia nigra and caudate nucleus. Dr. Raj and his team also found that the contributions of various brain regions to these symptoms changed over time, suggesting that the regions responsible for certain impairments shifts as the disease progresses. For instance, their model revealed that the red nucleus plays a primary role in cognitive scores early on but diminishes by the fourth year when the substantia nigra is most involved in cognitive impairments. As for motor scores, the palladium is most dominant in the second year and the role of the subcallosal area grows over time. These results offer new insights about PD progression and symptomology and highlight the growing importance of using ML methods as powerful diagnostic tools in medicine.