The article outlines the benefits and challenges of classifying neurons and highlights new genetic techniques for defining cell types.
In the August 7 issue of Science, science writer Emily Underwood elegantly lays out the historical background—dating back to the research of Santiago Ramón y Cajal—underlying The BRAIN Initiative’s goal of classifying the enormous variety of cells in the brain. Underwood goes on to spotlight a handful of new genetic techniques for cataloging neuronal diversity.
Underwood explains that while some neurological diseases, such as amyotrophic lateral sclerosis, are caused by disruptions within a single known cell type, other diseases, such as schizophrenia and autism, likely involve various cell types, some of which are, as yet, undocumented. Underwood writes that early classification schemes took note of neurons’ shapes and branching patterns, whereas mid-century neuroscientists used neurons’ firing patterns and the neurotransmitters they released to further split the cells into different groups. Modern genetic techniques, she states, are parsing neurons even more finely.
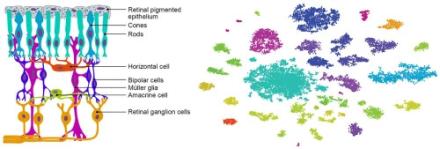
One of the new genetic methods that Underwood mentions, called Drop-Seq, is used by NIH BRAIN Initiative grantee Joshua Sanes, PhD, of Harvard University and colleagues. This technique has been developed in collaboration with Steven McCarroll and was described by lead author Evan Macosko, MD, PhD, and others in a recent issue of Cell. The approach uses a high throughput and cost-effective way to sort individual cells that are then classified based on the production of messenger RNA (mRNA), which reflects gene expression. See this post for more details.
Near the end of the article, Underwood conveys the thoughts of NIMH program director Andrea Beckel-Mitchener, PhD, co-leader of the team that manages NIH BRAIN Initiative grants on the cell census. According to Beckel-Mitchener, it will take multiple threads of data—including cell morphology, electrical activity, and gene activity—to reliably classifying neurons. This is consistent with conclusions drawn at a recent NIH-supported workshop where participants discussed how one might define cellular characteristics that help classify cell types in the brain.
