Microbubbles help lasers focus on neurons deep in the brain…New genetically encoded voltage indicator reveals individual action potentials…Scientists use RNA sequencing to classify neurons in primary visual cortex.
Microbubbles help lasers focus on neurons deep in the brain
Caltech scientists, led by BRAIN Initiative grantee Dr. Changhuei Yang, have designed a method for improving the focus of laser beams used to probe neurons in deep brain structures. Described in a recent issue of Nature Communications, the method, called time-reversed ultrasound microbubble encoded (TRUME) optical focusing, seeks to overcome the opacity of brain tissue, which can distort light, thereby making it difficult to image neural activity. TRUME uses ultrasound waves to destroy microbubbles embedded in neural tissue, increasing the precision of optical measurements of neural activity deep in the brain.

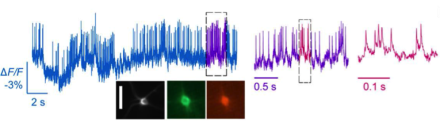
New genetically encoded voltage indicator reveals individual action potentials
Scientists led by BRAIN Initiative grantee Dr. Mark Schnitzer of Stanford University have developed a new technique for imaging electrical activity from individual neurons in the brains of mice and flies with a resolution of about 0.2 milliseconds. Capturing neural activity at such high speeds allows researchers to see not just action potentials, but also more subtle electrical activity in the dendrites of neurons that precede the firing of action potentials. The key to the new technique, reported recently in the journal Science, is a novel fluorescent marker that couples the fast voltage sensing domains of the A. acetabulum rhodopsin protein and a bright fluorophore.
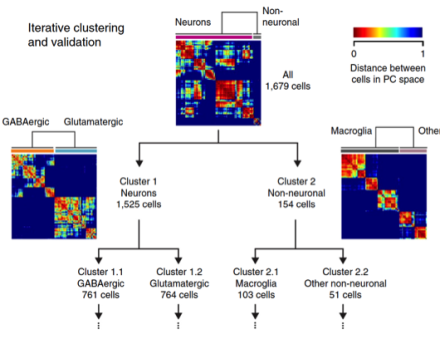
Scientists use RNA sequencing to classify neurons in primary visual cortex
BRAIN Initiative grantee Dr. Hongkui Zeng and colleagues at the Allen Institute for Brain Science, have used single-cell RNA sequencing to classify the cells of the primary visual cortex (area V1) of mice. Based on this transcriptomic analysis of more than 1,600 V1 cells, the team identified 49 cell types, including 23 that primarily release the brain chemical GABA, 19 that primarily release the brain chemical glutamate, and 7 non-neuronal cell types. These divisions include all the major V1 cell types reported in the literature, some additional new types, as well as subdivisions among the major types. Their analysis, reported recently in the journal Nature Neuroscience, also revealed that some of the cell types displayed unique electrophysiological and axon projection properties, thereby confirming that the single-cell transcriptomic signatures can be associated with specific cellular properties.